Modelling animal invasive species distribution
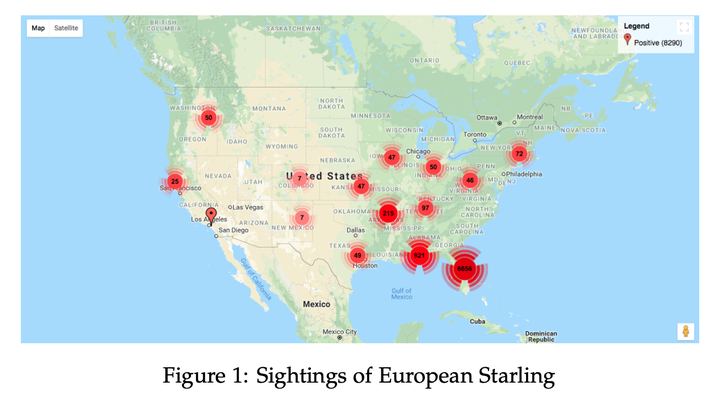
In the past few decades, a number of methods have been proposed to model species distributions yet there is no goto approach for applied researchers to model a given species. Given the publicly available datasets of detailed climatic and topological data along with the effort and collaboration of researchers to generate reliable location data of different species, predictive modelling of species distribution has become substantially feasible and reliable. Species distribution models have been applicable in many fields such as climate change biology, conservation biology, and landscape ecology. Although the type of data available for different species varies, there are usually two categories of data for a given species: detailed presence/absence data or presence-only data. Although presence/absence data is certainly more preferable as compared to presence only data, however, in a more practical scenario absence data is relatively more difficult to collect and for most species only the presence only data can be found. In this article, we attempt to provide some useful insight into the performance of standard predictive models such as simple models (decision trees, naive bayes), complex models (gradient boosting machines, neural networks) and popularly used models (Maxent). Additionally, we analyze the influence of using pseudo-negative samples to assess a model and propose an alternative approach based on generative adversarial networks to model species distribution using presence only data. Our empirical study, based on two invasive species namely European starling and zebra mussel supports the validity and effectiveness of our proposed approach.